HOMOLOGY RECOGNITION WITHOUT SEPARATING THE DOUBLE STRAND?
Recognition of a homologous sequence happens with quite the precision, and with complex molecular machinery, but to recognize a sequence without separating it from the double strand? Seems amazing right? Well, Researchers from Tokyo have discovered so. Homologous recognition is the basis of meiotic recombination for gamete formation in sexual reproduction and also acts as a DNA damage repair mechanism maintaining the stability of the genome, which is crucial for a cell's reproduction and survival.
 |
| Holliday model of recombination source: Fig. 30-67 from Biochemistry 4th ed., by Voet and Voet (John Wiley & Sons) |
 |
| Mechanism of Homology directed DNA repair source: https://www.nature.com/articles/cr20081 |
Interestingly, previous research had shown that only two RecA protomers were required for homology recognition to form the hybrid-duplex core. However, The filament structure provides multiple binding sites for the dsDNA and thus accelerates D-loop formation.
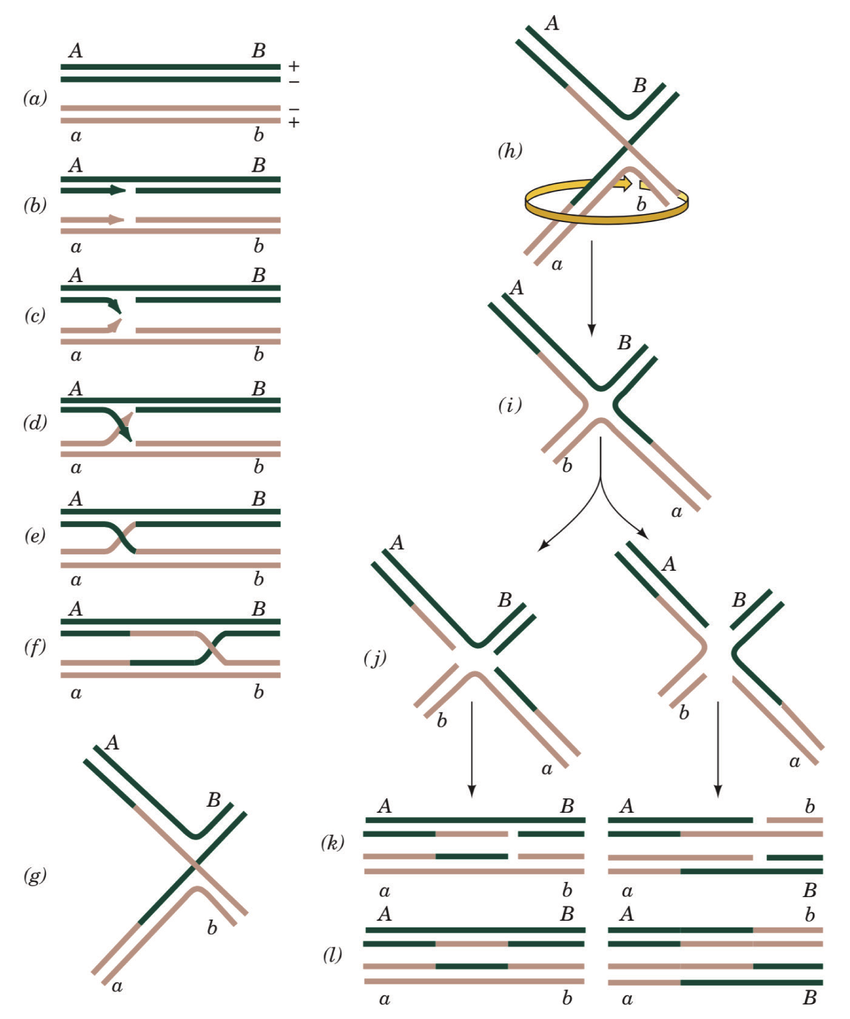
Two explanatory models have been proposed by researchers previously to explain the molecular mechanism behind homologous recognition: “ssDNA annealing after dsDNA-strand separation” and “homology recognition without dsDNA-strand separation”
The "ssDNA annealing after dsDNA-strand separation" model suggests that homology recognition occurs after the DNA strands have been separated, allowing the single-stranded DNA (ssDNA) to anneal with the complementary sequence in the double-stranded DNA (dsDNA). This model proposes a more straightforward mechanism where the ssDNA pairs with the dsDNA after the strands have been fully separated.
On the other hand, the "homology recognition without dsDNA-strand separation" model suggests that homology recognition occurs without complete separation of the DNA strands. Instead, this model proposes that the ssDNA recognizes and pairs with the complementary sequence in the dsDNA without fully separating the strands.
 |
| The two models for Homologous Recombination source: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkad1260/7517491 |
A recent study supporting the former theory explained that the phenylalanine at position 203 (Phe203) in the β-hairpin loop in eubacterial RecA stabilized the dsDNA separation by preventing the pairing up of the first-opened base pairs in the D-loop. The researchers of this study replaced the Phe203 with Alanine (F203A) and discovered that homology recognition indeed happens without separating the double strands thus supporting the latter theory. Although the mutant RecA lacked the amino acid to prevent the collapse of a double-strand separation, the hybrid-duplex product was formed! This meant that the dsDNA did not have to be separated for homology recognition. They also found that during the search for a homologous sequence in the complex, the dsDNA for approximately up to 3000 bp was not unwound by even a single turn!
The researchers of this study also state that although the former theory seems straightforward, the unwinding of dsDNA along with strand separation causes rotational stress on the DNA, especially in vivo and therefore the latter mechanism gives an advantage to the dsDNA to carry out the process quickly and much more efficiently.
So, the question now is, how does homology recognition happen without separating the double strand? The researchers explained in the study that dsDNA separates orderly at a specific temperature but at the physiological temperature, each of the base pairs opens and pairs randomly wherein the lives of G-C pairs last longer than that of A-T pairs. Furthermore, when the base stacking is disturbed under certain environments such as local expansion between base pairs due to thermal molecular movements or the bending of the dsDNA, the unpaired bases rotate and get exposed outside the helix. These exposed bases form Watson - Crick type of pairs with the ssDNA for homology recognition by RecA without separating the strand.
REFERENCE:
Shibata T, Ikawa S, Iwasaki W, Sasanuma H, Masai H, Hirota K. Homology recognition without double-stranded DNA-strand separation in D-loop formation by RecA. Nucleic Acids Research. 2024 Jan 12:gkad1260.


Comments
Post a Comment