FORGING CELLULAR ALLIANCES: A LEAP TOWARD UNDERSTANDING ENDOSYMBIOSIS
Endosymbiosis is the phenomenon where one organism called endosymbiont resides within another organism called host. It is considered as the major force behind eukaryotic cell evolution. These symbiotic relations may range from antagonist to mutually beneficial relationships. The significant endosymbiosis happened ∼1.8 billion years ago when Asgard archean formed an endosymbiotic relationship with an alphaproteobacterium which gave mitochondria. This greatly expands the energy production and expenditure of single and multicellular eukaryotes playing a vital role in cell cycle regulation, signaling and apoptosis and forms the basis for further endosymbiotic relationships. Soon after eukaryogenesis at about 1.5 billion years ago an ancestor to Archaeplastida (algae and plants) established another significant endosymbiosis with cyanobacterium which results in establishment and evolution of chloroplasts leading to photosynthesis and massively increasing the global productivity.
 |
| Major endosymbiosis events |
Apart from these major endosymbiotic events, different eukaryotic lineages acquired further endosymbiosis which provided novel metabolic pathways and enhanced nutrient utilization to the host. These are seen from single-celled eukaryotes to plants and invertebrates. One such example is association of plants with arbuscular mycorrhizae, nitrogen-fixing cyanobacteria, and rhizobia bacteria. These events are important for early success of land plants and further diversification. A wide range of marine invertebrates formed endosymbiosis with chemosynthetic bacteria for CO2 fixation to form consumable biomass and sugars. Endosymbiosis is responsible for 20% of insect diversity which accounts for >1 million species
However, the phenomenon of endosymbiosis is rare due to the fact that endosymbionts must first enter the host cell, and overcome challenges associated with immune responses, metabolism and growth synchronization. Thus early stages in endosymbiosis remained elusive and primarily studies had been conducted on insects which had revealed important aspects of endosymbiosis such as control of bacterial replication. But attempts in other organisms have not resulted in stable endosymbiosis. Recently scientists developed a novel endosymbiotic relationship between early divergent fungus Rhizopus microsporus and cytosolic bacterial endosymbiont Mycetohabitans rhizoxinica such that fungus cannot sporulate without the endosymbiont and endosymbiont colonizes the spores allowing for a strict vertical inheritance, in order to study the early endosymbiogenesis events in detail.
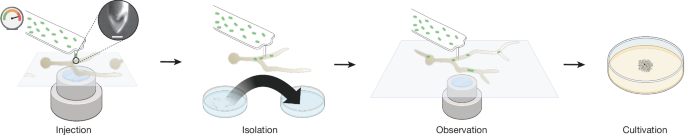 |
| Workflow of injection of bacteria in fungal hyphae to develop novel endosymbiosis |
The bacteria was injected in the fungal hyphae using Fluidic force microscopy (FluidFM) using specially designed double point cylindrical tip, whose tip made using focused ion beam milling and has an aperture of 50-1000 nm to allow passage of bacteria with minimal damage and this technique allowed the injection of 1-30 bacteria per injection event into R.microsporus germling whose cell wall was previously softened using an enzyme mixture in osmoprotective solution. The colonization was confirmed by presence of green fluorescent protein (GFP) labelled M.rhizoxinica using microscopy. On the basis of the velocity, bidirectionality, and the path followed with respect to movement of general cytoplasmic contents, the endosymbiont showed both active transport along the microtubules and passive transport through cytoplasmic bulk flow. Active transport along the microtubules was suspected to be mediated by M.rhizoxinica attachment to microtubule-dependent dyneins and kinesins, or through cargo adaptor proteins. To test whether this is the case, the function of dyneins were inhibited by ciliobrevin D and a transient halt of bacterial transport within the germling. The stable vertical inheritance of endosymbiont was confirmed using Fluorescence-activated cell sorting (FACS) analysis of a large number of spores.
Injection of E.coli in R.microsporus was followed by rapid replication of the bacterium within the hyphae followed by the clumping of the bacterium which were slowly dispersed along the cytoplasmic bulk flow. These clumps got localized and these highly populated hotspots were separated by the formation of septum, the compartment without E.coli continues to grow normally but the compartment with E.coli gets completely filled and bursts open. The septa formation was also observed in the injection of M.rhizoxinica mutants lacking the effector Mycetohabitans transcription activator-like effector 1 (MTAL1) which helps the bacteria to evade the fungal defences. The FACS analysis revealed the absence of E.coli colonization of spores thereby disproving the stable inheritance of the endosymbiont. This showed that fungus had recognized the unadapted bacterium and produced the defence response to physically contain the bacterium and allowing the normal growth of uninfected hyphae.
 |
| a) Injection of E.coli which is followed by rapid reproduction, localization and septum formation. b) Injection of M.rhizoxinica showing its replication and movement in fungal hyphae |
FACS analysis revealed a subset of spores which were colonized by the bacteria and then grown on a rich medium; these positively sorted spores germinated successfully and retained a high population of replicating bacteria that spread throughout the hyphae.
| Germling from positively sorted spores with endosymbiont distributed throughout the germling |
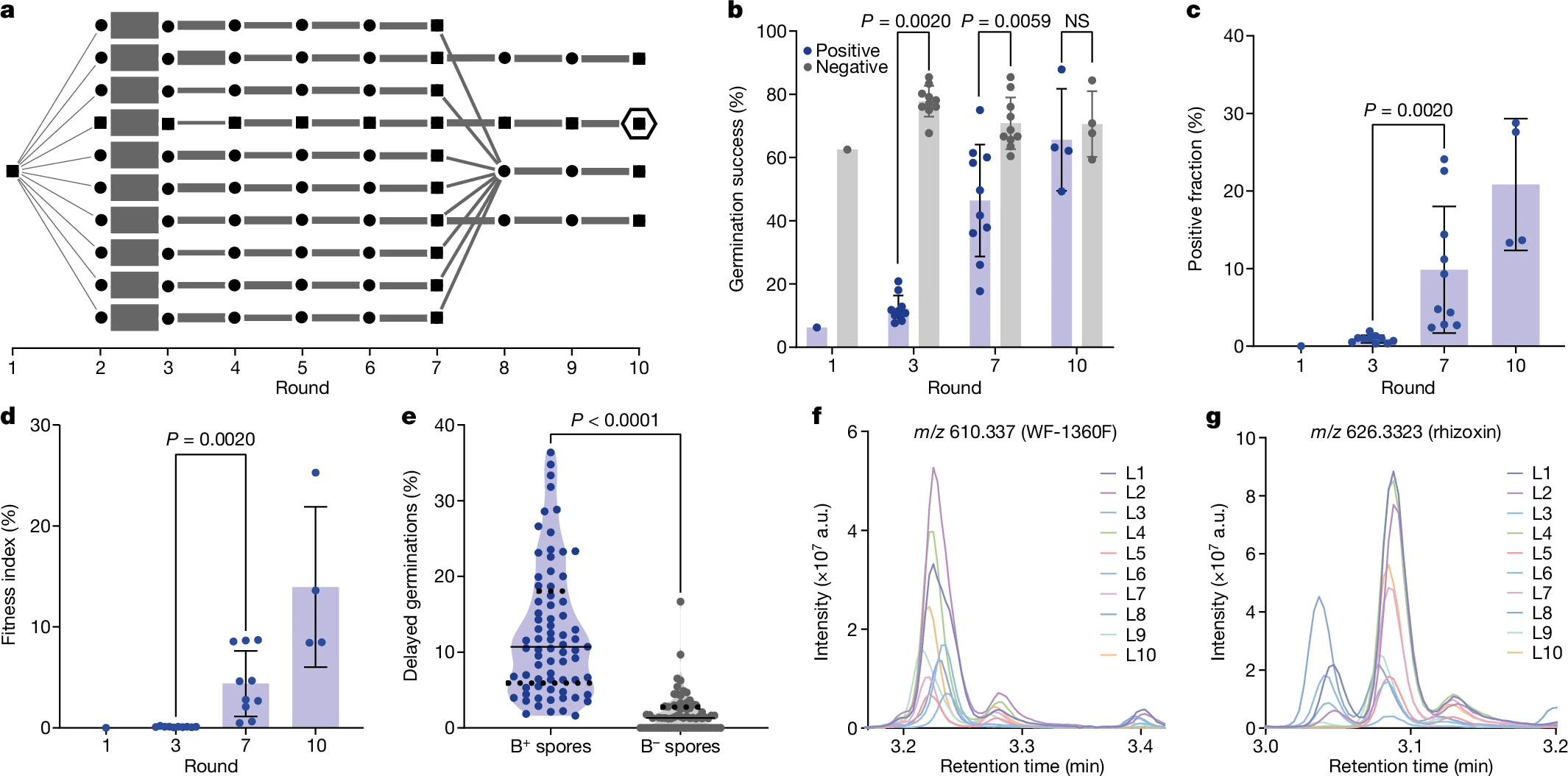
Initially, the germination success of spores not containing bacteria was around 63% but in the presence of the bacteria it was reduced tenfold to 6.3% indicating the substantial cost posed by the bacterium on the fungi. But following several rounds of replication and selection of to get high performing germlings with the germination success rate of 75% following tenth round of selection. In the same way the positive fraction had increased from 0.01% to 24.1%. These results increased the fitness index of the endosymbiosis from initial 0.0006% to 8.7% with an average of 4.4% after round 7 and after round 10 it had reached 14% with a highest value of 25.3%. Additional cost to fungus was observed in the form of delayed germination of spores with bacteria that do not contain the bacteria.
To test whether introduction of the endosymbiont had lead to transfer of biosynthesis of natural projects, analogous to the natural endosymbiosis relationships, the production of macrocyclic polyketides such as WF-1360F by M. rhizoxinica and subsequent conversion to rhizoxins and other congeners by R. microsporus was tested using liquid chromatography with tandem mass spectroscopy. R. microsporus is resistant to action of rhizoxin due to single amino acid substitution in β-tubulin preventing its binding and subsequent prevention of inhibition of microtubule formation. The liquid chromatography with tandem mass spectroscopy analysis revealed the presence of WF-1360F and rhizoxin on plates of ten evolution lines in round 7 which validated the transfer of metabolic trait to the host which do not generate the natural product on its own.
It was observed the bacterial load is negatively correlated to the germination success at the initial ancestral pair with spores with lower bacterial load had a higher germination success but in evolved pair all spores had relatively low bacterial load compared to ancestral pair and these suggests that the endosymbiosis had adapted to reduce the bacterial load to circumvent the issue of negative correlation of bacterial load with germination success. It was also observed that germination success of both ancestral and evolved pairs continuously decreased from 50% to 5% after a month suggesting the presence of bacteria takes continuous toll on the spores.
| Germination success of bacteria-positive spores (Pos) decreases faster than bacteria-negative spores (Neg) |
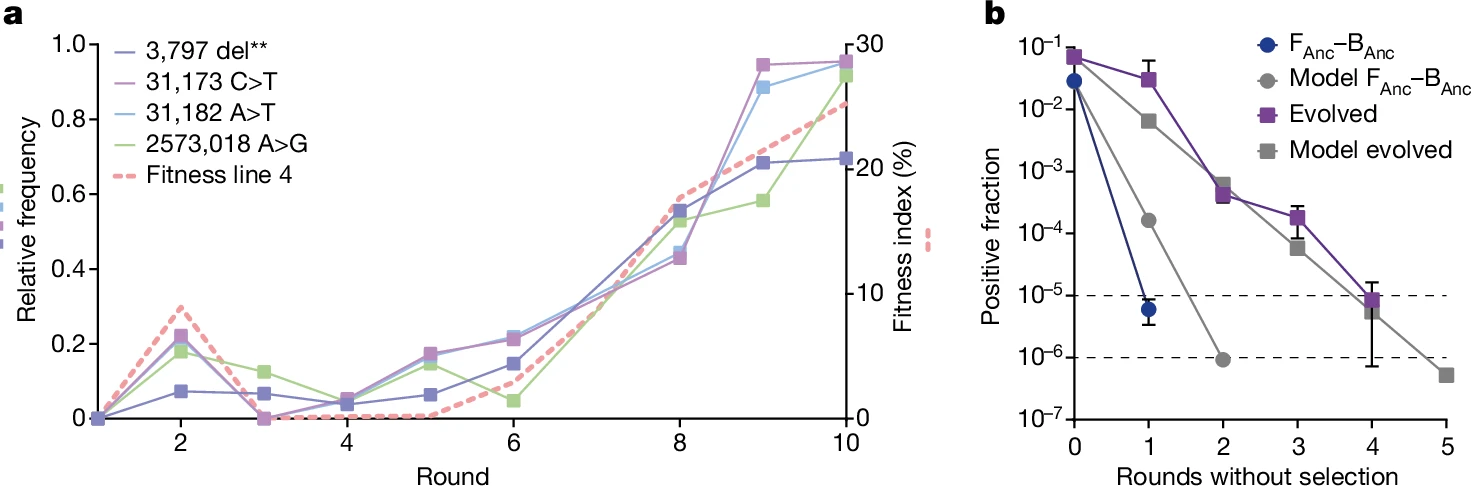
The presence of genetic changes was tested and it was found that the endosymbiont didn't develop any mutational changes throughout the experiment but the fungal host had developed nine mutations which analysis using InterPro and STRING revealed that they may be linked to endocytosis, translation, mitochondrial maintenance and ion transport suggesting that genetic changes happen at the host side rather than in endosymbiont side. The increase in prevalence of mutations was found to be positively correlated to fitness index. To test the stability of the endosymbiosis in the absence of positive selection, a mathematical model based on germination success and initial bacterial colonization was used to predict how the positive fraction would change over time. In the ancestral group, the positive fraction decreased very quickly, dropping below threshold of propagation (<1/100,000) but still in detectable range (>1/1,000,000) after just one round, indicating that endosymbiosis was not stable at early stages. However, in the evolved group, the positive fraction declined more slowly over 4-5 rounds, aligning with the model's predictions. This result suggests that adaptive evolution improved the stability of the endosymbiosis.
In conclusion, endosymbiosis has long been recognized as a fundamental force driving evolutionary complexity, yet the early stages of its formation remain a mystery. This groundbreaking research on inducing endosymbiosis in fungi not only provides a rare glimpse into the origins of these cellular partnerships but also opens new avenues for synthetic biology and evolutionary studies. By engineering stable endosymbiotic relationships, scientists can explore how cooperation between species evolves, how metabolic traits are transferred, and how life itself adapts to new symbiotic paradigms.
REFERENCES
Gordon M Bennett, Younghwan Kwak, Reo Maynard, Endosymbioses Have Shaped the Evolution of Biological Diversity and Complexity Time and Time Again, Genome Biology and Evolution, Volume 16, Issue 6, June 2024, evae112, https://doi.org/10.1093/gbe/evae112
Giger, G.H., Ernst, C., Richter, I. et al. Inducing novel endosymbioses by implanting bacteria in fungi. Nature 635, 415–422 (2024). https://doi.org/10.1038/s41586-024-08010-x
IMAGE SOURCE

Comments
Post a Comment